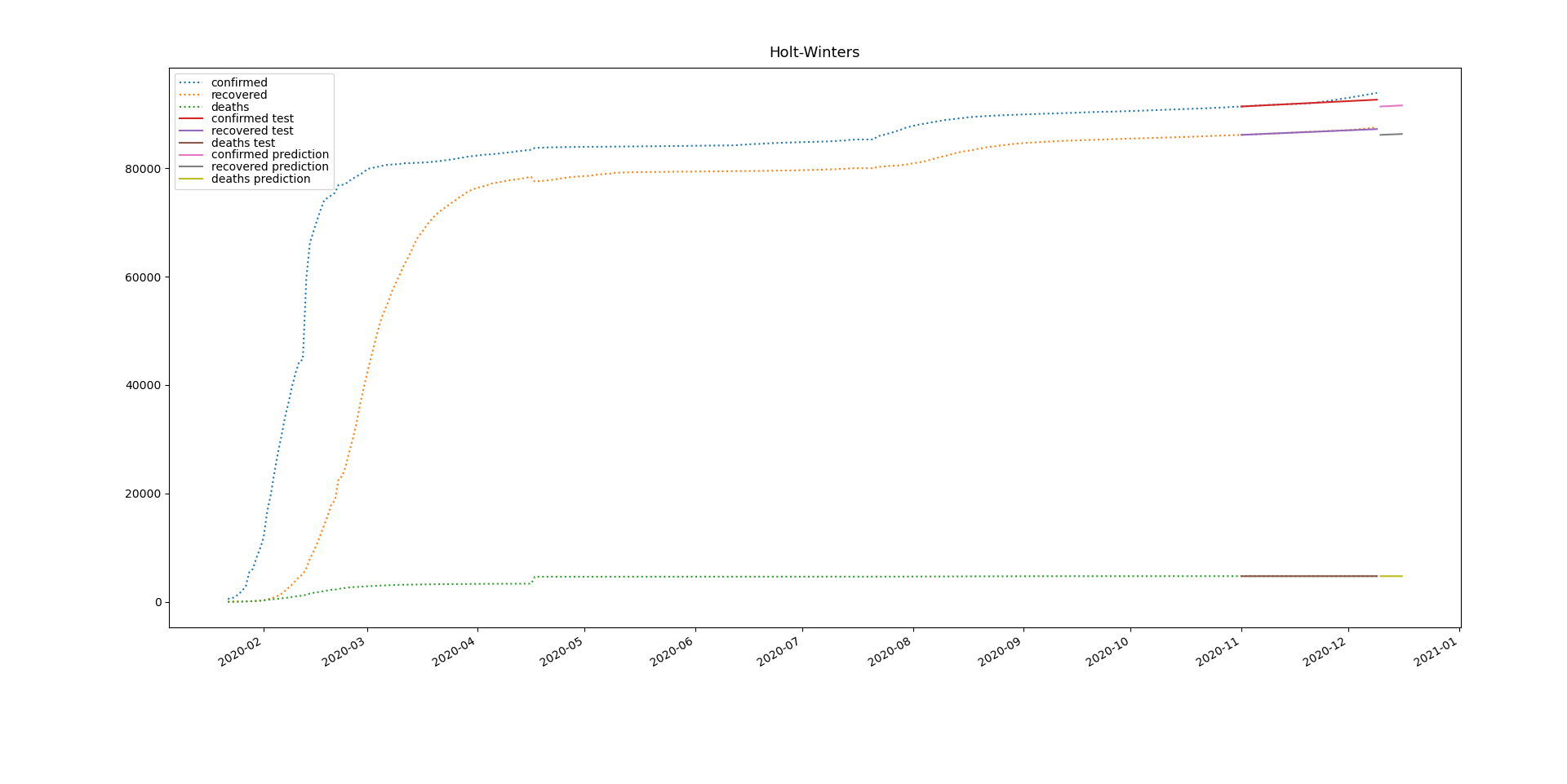
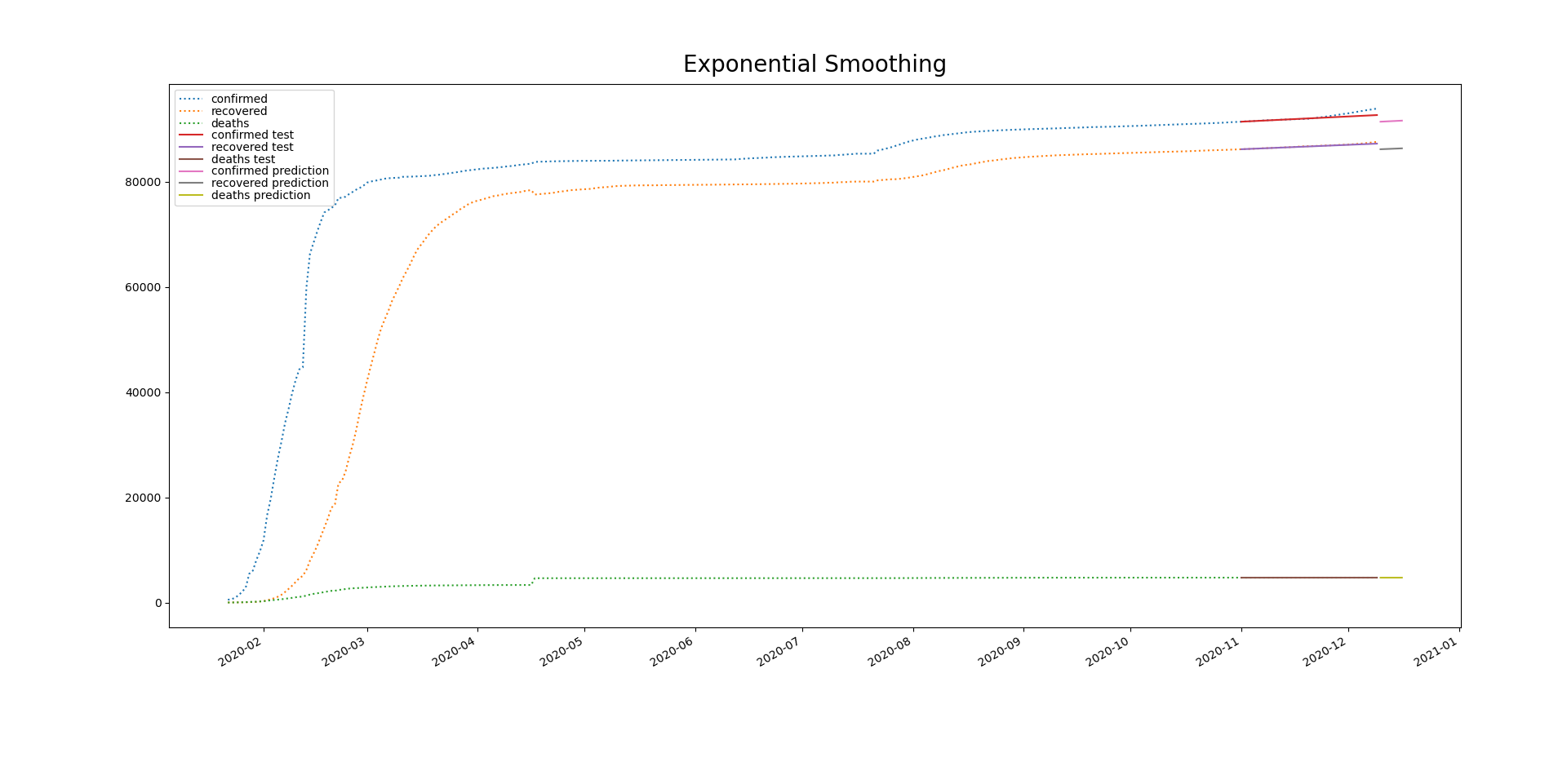
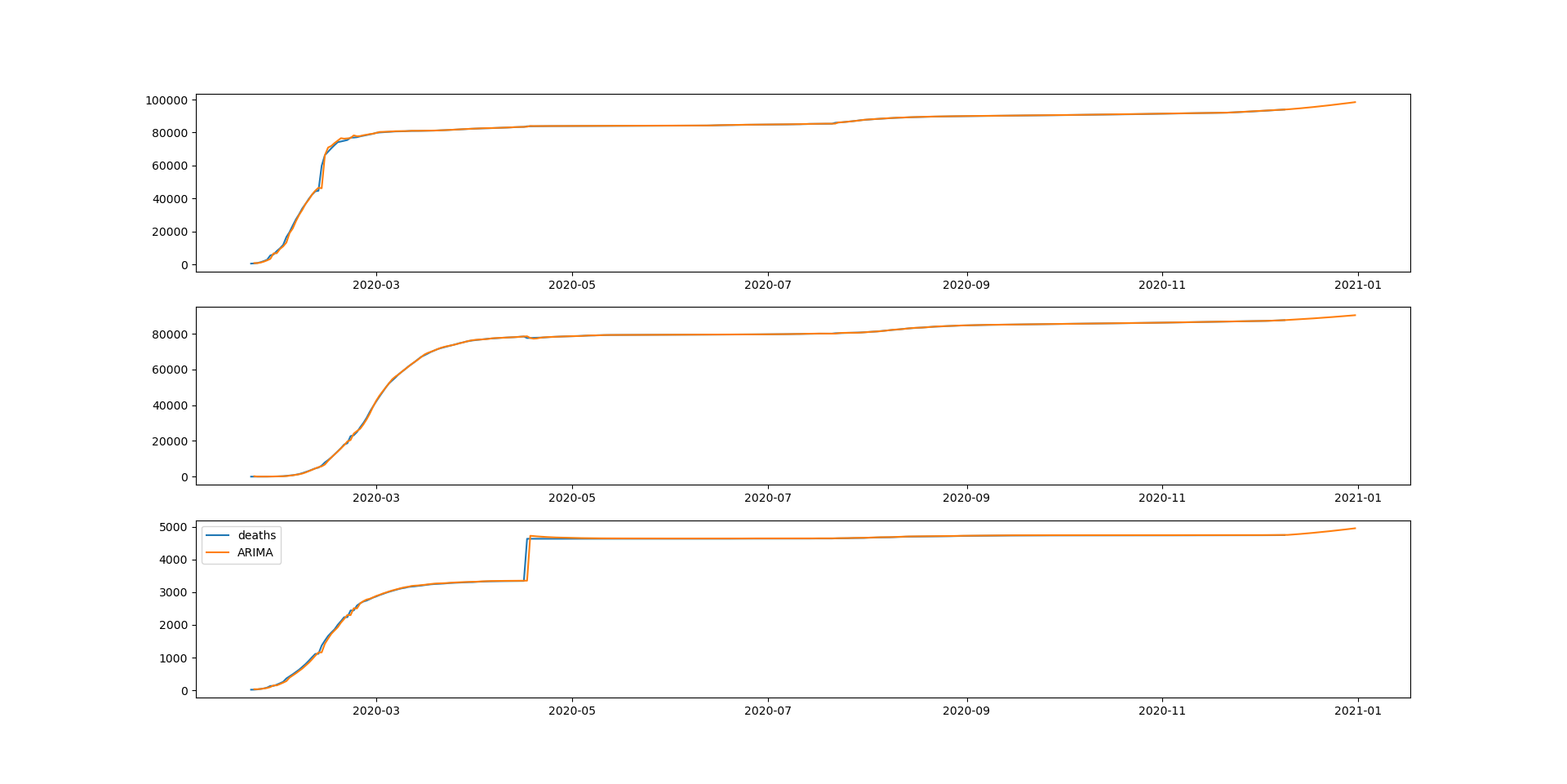
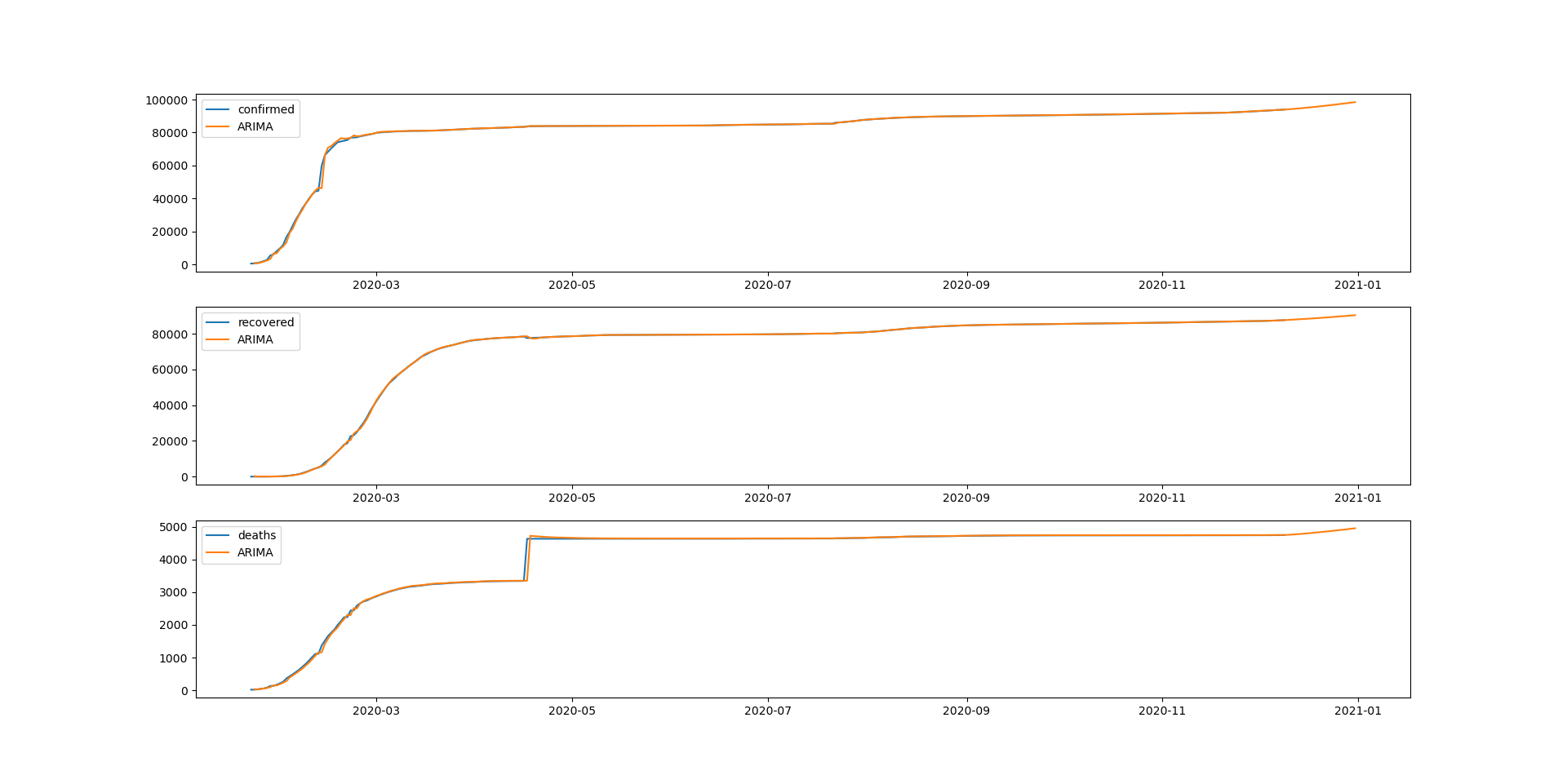
19 changed files with 9 additions and 7 deletions
BIN
COVID-19/Prediction/.vs/Prediction/v16/.suo
View File
+ 3
- 1
COVID-19/Prediction/Prediction/CN.py
View File
+ 1
- 1
COVID-19/Prediction/Prediction/CNConPara.py
View File
+ 1
- 1
COVID-19/Prediction/Prediction/CNDeaPara .py
View File
+ 1
- 1
COVID-19/Prediction/Prediction/CNRecPara.py
View File
+ 3
- 3
COVID-19/Prediction/Prediction/ExponentialSmoothing.py
View File
BIN
COVID-19/Visualization/.vs/Visualization/v16/.suo
View File
BIN
Essay/COVID-19.pptx
View File
BIN
Essay/基于新冠疫情数据的分析与建模.docx
View File
BIN
Essay/基于新冠疫情数据的分析与建模.pdf
View File
BIN
Essay/基于新冠疫情数据的分析与建模.pptx
View File
BIN
ParaCon/Figure_1.png
View File
BIN
ParaCon/Figure_2.png
View File
BIN
ParaCon/Figure_3.png
View File
BIN
ParaDea/Figure_1.png
View File
BIN
ParaRec/Figure_1.png
View File
BIN
ParaRec/Figure_2.png
View File
BIN
prediction_figure/Figure_1.png
View File
BIN
prediction_figure/Figure_4.png
View File
Loading…